Research
Papers
Posters
Collaborations
Visiting students
Links
Mirrored tools
SMS2
Vintage science
Overview
21/01/2021
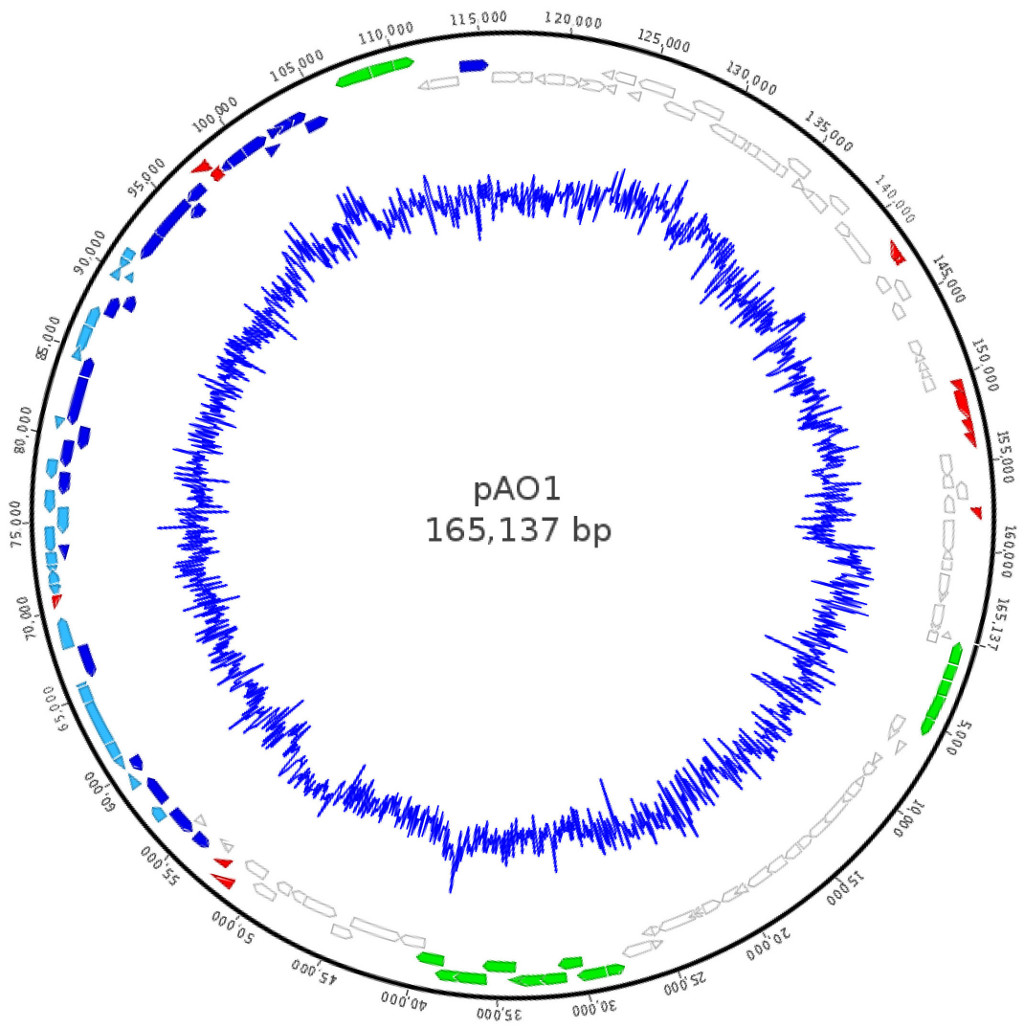
Paenarthrobacter nicotinovorans (formerly Arthrobacter nicotinovorans, Arthrobacter oxidans) is a coryneform bacteria able to grow on nicotine as sole carbon and nitrogen source due to the presence of the 165 kb megaplasmid pAO1. The megaplasmid was sequenced and contains 2 major gene clusters: one related to nicotine metabolism and a second cluster forming an operon related to sugar catabolism.
The
pAO1 catabolic megaplasmid of Paenarthrobacter nicotinovorans (shades
of blue - genes related to nicotine uptake and utilization,
green - xylose catabolism related genes, red - recombination
related genes, white - unknown function)
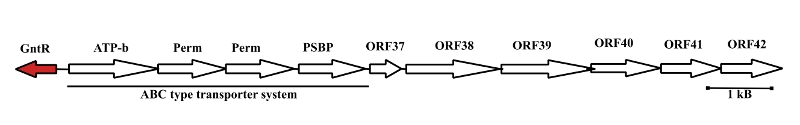
I am mainly focused on the molecular biology of the pAO1 megaplasmid related to the defense mechanism against nicotine-generated oxidative stress, sugar-catabolism and biotechnological applications.
Using a unique
combination of BLAST searches, protein modeling and in-silico
docking I identified putative substrates for the pAO1 coded
sugar-operon. The selected compounds were experimentally tested
and xylose was identified as the physiological substrate.
The
operon coding for a oxidative xylose-catabolic pathway on
the pAO1 megaplasmid.
In the last
couple of years I
focused on the possible
medical applications of the nicotine-derivatives produced by Paenarthrobacter
nicotinovorans.
Using in-silico docking experiments, we tested the
potential of nicotine and piridine derivatives produces by
this bacteria to bind nAchR and identified 6-Hydroxy-Nicotine
(6HNic) as a potential ligand. In
collaboration with dr.
Lucian Hritcu we showed
that indeed 6HNic is
able to modulate the function of nAChRs and sustain
spatial memory formation in a rat model of Alzheimer’s
disease. Currently we are working on a biotechnological
method to produce and isolate 6HNic using a genetically
engineered Paenarthrobacter nicotinovorans
strain.



Growing Paenarthrobacter
nicotinovorans
in a 3L micro-fermenter.
48 hour of growth accelerated in 1 min and 30 secs thanks to PhD Eugen Ungureanu
The latest projects involves the proteomics, transcriptomics and genomic analysis of Paenarthrobacter nicotinovorans pAO1 and its nicotine degradation pathway. Till now, the lab has produced several relevant datasets:
-
the first complete and annotated genome of an nicotine-degrading bacteria Paenarthrobacter nicotinovorans ATCC 49199: GenBank CP089293 and CP089294, as well as the associated raw data: PRJNA693273
-
a transcriptomics data-set dealing with the nicotine metabolism in Paenarthrobacter nicotinovorans ATCC 49199: Gene Expression Omnibus GSE240220
-
a shotgun proteomics dataset that identifies the proteins expressed by Paenarthrobacter nicotinovorans in the presence of nicotine by using nanoLCMSMS PXD008756
-
another shotgun proteomics dataset that performs a time-based analysis of the nicotine catabolism in Paenarthrobacter nicotinovorans - PXD012577
Funding
-
PN-III-P4-ID-PCE-2020-0656, Sequencing the genome of a useful bacteria: Paenarthrobacter nicotinovorans – next step in extending it’s biotechnological applications
-
PN-III-P2-2.1-TE-2016-0367 - Developing an Arthrobacter nicotinovorans biotechnology for neuro-pharmaceuticals production
-
PN-III-P2-2.1-PED-2016-0177 - Steps towards an Arthrobacter nicotinovorans biotechnology for neuro-pharmaceuticals production
-
PN-II 50BM/2016 - Romania - P.R.P. China joint research project - Nicotine - from toxic residue to metabolic derivatives with neuroprotective effects
-
PN-II-RU-TE-2014-4-0106 – Effects of 6-hydroxy-nicotine on chlorisondamine-induced oxidative stress and neurotoxicity: relevance for Alzheimer’s disease
-
GI-2014-02 – Internal Research grant awarded by the Alexandru I Cuza University of Ia?i
-
TD-236 research grant, Molecular implications of the pAO1 megaplasmid for the Arthrobacter nicotinovorans
-
PD-337 research grant, Cloning and characterization of ORF32 and ORF40 from the pAO1 megaplasmid of Arthrobacter nicotinovorans -potential protein-tagatose interactions models
Selected publications
Papers/Chapters
published in the last 5 years
- El-Sabeh, A., Mlesnita, A. M., & Mihasan, M (2025). Integrated transcriptomic and proteomic analysis of nicotine metabolism in Paenarthrobacter nicotinovorans ATCC 49919. International Biodeterioration & Biodegradation, 199, 106017. [Free Fulltext]
- Răzvan-Ștefan,
B., Laura
Nicoleta, P.,
& Mihasan,
M.
(2025). Impact
of 3D-printed
molecular
models on
student
understanding
of
macromolecular
structures: a
compensatory
research
study.
Biochemistry
and Molecular
Biology
Education. [Fulltext]
- Mantea, L.-E.; El-Sabeh, A.; Mihasan, M.; Stefan, M. Bacillus safensis P1.5S Exhibits Phosphorus-Solubilizing Activity Under Abiotic Stress. Horticulturae 2025, 11, 388. [Fulltext]
- Postu, P.A.; Boiangiu, R.S.; Mihasan, M.; Stache, A.B.; Tiron, A.; Hritcu, L. The Distinct Biological Effects of 6-Hydroxy-L-Nicotine in Representative Cancer Cell Lines. Molecules 2024, 29, 5593. [Free Fulltext]
- Brinza,
I., Boiangiu,
R. S.,
Mihasan, M.,
Gorgan, D. L.,
Stache, A. B.,
Abd-Alkhalek,
A., El-Nashar,
H., Ayoub, I.,
Mostafa, N.,
Eldahshan, O.,
Singab, A. N.,
& Hritcu,
L. (2024).
Rhoifolin,
baicalein
5,6-dimethyl
ether and
agathisflavone
prevent
amnesia
induced in
scopolamine
zebrafish
(Danio rerio)
model by
increasing the
mRNA
expression of
bdnf, npy,
egr-1, nfr2α,
and creb1
genes. European
Journal of
Pharmacology,
984, 177013
[Free
Fulltext]
- Boiangiu,
R.S.; Brinza,
I.; Honceriu,
I.; Mihasan,
M.;
Hritcu, L.
Insights into
Pharmacological
Activities of
Nicotine and
6-Hydroxy-L-nicotine,
a Bacterial
Nicotine
Derivative: A
Systematic
Review.
Biomolecules
2024, 14, 23.
[Free
FullText]
- El-Sabeh,
A., Mlesnita,
AM., Munteanu,
IT. .... Mihasan
M.
Characterisation
of the
Paenarthrobacter
nicotinovorans
ATCC 49919
genome and
identification
of several
strains
harbouring a
highly
syntenic
nic-genes
cluster. BMC
Genomics 24,
536 (2023). [Free
Fulltext]
- Pérez-Ramos,
A.; Ladjouzi,
R.; Mihasan,
M.; Teiar,
R.; Benachour,
A.; Drider, D.
Advances in
Characterizing
the Transport
Systems of and
Resistance to
EntDD14, A
Leaderless
Two-Peptide
Bacteriocin
with Potent
Inhibitory
Activity. Int.
J. Mol.
Sci. 2023,
24, 1517. [Free
Fulltext]
- Amada El-Sabeh, Iasmina Honceriu, Fakhri Kallabi, Razvan-Stefan Boiangiu, Marius Mihasan - Complete Genome Sequences of Two Closely Related Paenarthrobacter nicotinovorans Strains. Microbiol Resour Announc. 2022; 11(6):e0013322. doi: 10.1128/mra.00133-22. [Fulltext]
- Madi-Moussa,
D.; Deracinois, B.; Teiar, R.;
Li, Y.; Mihasan, M.;
Flahaut, C.; Rebuffat, S.;
Coucheney, F.; Drider, D.
Structure of Lacticaseicin 30
and Its Engineered Variants
Revealed an Interplay between
the N-Terminal and C-Terminal
Regions in the Activity against
Gram-Negative Bacteria. Pharmaceutics
2022, 14, 1921.
doi:10.3390/pharmaceutics14091921 [Fulltext] - Postu, P.A.; Mihasan, M.; Gorgan, D.L.; Sadiki, F.Z.; El Idrissi, M.; Hritcu, L. Pinus halepensis Essential Oil Ameliorates Aβ1-42-Induced Brain Injury by Diminishing Anxiety, Oxidative Stress, and Neuroinflammation in Rats. Biomedicines 2022, 10, 2300. doi:10.3390/biomedicines10092300 [Fulltext]
- Mihasan M., Boiangiu R.S., Guzun D., Babii C., Aslebagh R., Channaveerappa D., Dupree D., Darie CC. Time-dependent analysis of Paenarthrobacter nicotinovorans pAO1 nicotine-related proteome. ACS Omega 2021, 6, 22, 14242–14251 [Fulltext]
- Mihasan,
M. A Beginner’s Guideline
for Low?cost 3D Printing of
Macromolecules Usable for
Teaching and Demonstration.
Biochem. Mol. Biol. Educ. 2021,
bmb.21493. [Fulltext]
- Postu,
P. A.; Tiron, A.; Tiron, C. E.;
Gorgan, D. L.; Mihasan, M.;
Hritcu, L. Conifer Essential
Oils Reversed Amyloid Beta1-42
Action by Modulating BDNF and
ARC Expression in The Rat
Hippocampus. CNS Neurol. Disord.
- Drug Targets 2021, 20. [Fulltext]
- Boiangiu,
R. S.; Mihasan, M.;
Gorgan, D. L.; Stache, B. A.;
Hritcu, L. Anxiolytic,
Promnesic,
Anti-Acetylcholinesterase and
Antioxidant Effects of Cotinine
and 6-Hydroxy-L-Nicotine in
Scopolamine-Induced Zebrafish
(Danio Rerio) Model of
Alzheimer’s Disease.
Antioxidants 2021, 10 (2), 212.
[Free
Fulltext]
- Brandsch, R and Mihasan M, A soil bacterial catabolic pathway on the move: Transfer of nicotine catabolic genes between Arthrobacter genus megaplasmids and invasion by mobile elements, Journal of Biosciences, 2020, 45, 1-12 [Fulltext]
- Djeussi,
D. E.; Noumedem, J. A. K.; Mihasan,
M.; Kuiate, J. R.; Kuete,
V. Antioxidant Activities of
Methanol Extracts of Thirteen
Cameroonian Antibacterial
Dietary Plants. J. Chem. 2020,
2020. [Free
Fulltext]
-
Dupree EJ,
Jayathirtha M, Yorkey H, Mihasan
M,
Petre BA,
Darie CC,
A Critical Review of Bottom-Up
Proteomics: The Good, the Bad,
and the Future of this Field,
Proteomes, 2020, 8 (3), 14 [Fulltext]
- Boiangiu
RS,
Mihasan M, Gorgan DL,
Stache BA, Petre BA, Hritcu L.,
Cotinine and
6-Hydroxy-L-Nicotine Reverses
Memory Deficits and Reduces
Oxidative Stress in
A?25-35-Induced Rat Model of
Alzheimer’s Disease,
Antioxidants, 2020, 9(8), 768 [Fulltext]
- Mih??an
M, Babii C, Aslebagh R,
Channaveerappa D, Dupree EJ,
Darie CC. Exploration of
Nicotine Metabolism in
Paenarthrobacter nicotinovorans
pAO1 by Microbial Proteomics.
In: Woods AG, Darie CC, editors.
Advancements of Mass
Spectrometry in Biomedical
Research, vol. 1140 of Advances
in Experimental Medicine and
Biology. 2019. p. 515–29. [Fulltext]
- Mih??an M, Wormwood KL, Sokolowska I, Roy U, Woods AG, Darie CC. Mass Spectrometry- and Computational Structural Biology-Based Investigation of Proteins and Peptides. In: Woods AG, Darie CC, editors. Advancements of Mass Spectrometry in Biomedical Research, vol. 1140 of Advances in Experimental Medicine and Biology. 2019. p. 265–87.[Fulltext]
The full list of publications is available here